Clustering and Dimensionality Reduction¶
Let's start with a classical example: IRIS dataset¶
The IRIS dataset contains measurements of 150 iris flowers from 3 species:
- Setosa
- Versicolor
- Virginica
Each flower has 4 features:
- Sepal length (cm)
- Sepal width (cm)
- Petal length (cm)
- Petal width (cm)
In [153]:
import pandas as pd
from sklearn.datasets import load_iris
# Load IRIS dataset
iris = load_iris()
In [154]:
X = iris.data # object features
y = iris.target # object labels
feature_names = iris.feature_names
target_names = iris.target_names
In [156]:
print(X[:5,:]) #print first 5 rows of X
print(y) # print object labels
[[5.1 3.5 1.4 0.2] [4.9 3. 1.4 0.2] [4.7 3.2 1.3 0.2] [4.6 3.1 1.5 0.2] [5. 3.6 1.4 0.2]] [0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2]
In [157]:
print(X.shape)
print(len(y))
print(feature_names)
print(target_names)
(150, 4) 150 ['sepal length (cm)', 'sepal width (cm)', 'petal length (cm)', 'petal width (cm)'] ['setosa' 'versicolor' 'virginica']
In [159]:
# Create a DataFrame for easier manipulation
df = pd.DataFrame(X, columns=feature_names)
df['species'] = pd.Categorical.from_codes(y, target_names)
In [89]:
print(df.head())
sepal length (cm) sepal width (cm) petal length (cm) petal width (cm) \ 0 5.1 3.5 1.4 0.2 1 4.9 3.0 1.4 0.2 2 4.7 3.2 1.3 0.2 3 4.6 3.1 1.5 0.2 4 5.0 3.6 1.4 0.2 species 0 setosa 1 setosa 2 setosa 3 setosa 4 setosa
In [160]:
print(pd.Categorical.from_codes(y, target_names))
['setosa', 'setosa', 'setosa', 'setosa', 'setosa', ..., 'virginica', 'virginica', 'virginica', 'virginica', 'virginica'] Length: 150 Categories (3, object): ['setosa', 'versicolor', 'virginica']
In [118]:
import seaborn as sns
from matplotlib import pyplot as plt
fig1 = plt.figure(figsize=(5, 5))
sns.pairplot(df)
Out[118]:
<seaborn.axisgrid.PairGrid at 0x7bd2c87822c0>
<Figure size 500x500 with 0 Axes>
In [161]:
from sklearn.cluster import KMeans
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
kmeans = KMeans(n_clusters=3, init='random', n_init=10, random_state=42)
cluster_labels = kmeans.fit_predict(X_scaled)
In [162]:
cluster_labels_category = pd.Categorical.from_codes(cluster_labels, target_names)
df['cluster'] = cluster_labels_category
In [163]:
sns.pairplot(df,hue='cluster')
Out[163]:
<seaborn.axisgrid.PairGrid at 0x7bd2b42468b0>
In [164]:
sns.pairplot(df,hue='species')
Out[164]:
<seaborn.axisgrid.PairGrid at 0x7bd2b4247ce0>
In [119]:
from sklearn.metrics import confusion_matrix
conf_matrix = confusion_matrix(y, cluster_labels)
print("Confusion Matrix:")
print("(Rows = True Species, Columns = Predicted Clusters)\n")
conf_df = pd.DataFrame(conf_matrix,
index=target_names,
columns=[f'Cluster {i}' for i in range(3)])
print(conf_df)
Confusion Matrix:
(Rows = True Species, Columns = Predicted Clusters)
Cluster 0 Cluster 1 Cluster 2
setosa 50 0 0
versicolor 0 38 12
virginica 0 14 36
Principal component analysis finds an orthogonal basis that best represents the variance in the data.¶

In [170]:
from sklearn.decomposition import PCA
pca = PCA(n_components=2);
X_pca = pca.fit_transform(X_scaled)
#Get cluster centers in PCA
centers_pca = pca.transform(kmeans.cluster_centers_)
In [165]:
fig, axes = plt.subplots(1,2, figsize = (16,5))
scatter1 = axes[0].scatter(X_pca[:,0],X_pca[:,1], c=y, cmap='Set1', s=100, alpha=0.6, edgecolors='black')
axes[0].set_xlabel(f'PC1 ({pca.explained_variance_ratio_[0]*100:.1f}% variance)', fontsize=12)
axes[0].set_ylabel(f'PC2 ({pca.explained_variance_ratio_[1]*100:.1f}% variance)', fontsize=12)
axes[0].set_title('Truth', fontsize=14, fontweight='bold')
scatter2 = axes[1].scatter(X_pca[:,0],X_pca[:,1], c=cluster_labels, cmap='Set1', s=100, alpha=0.6, edgecolors='black')
axes[1].set_xlabel(f'PC1 ({pca.explained_variance_ratio_[0]*100:.1f}% variance)', fontsize=12)
axes[1].set_ylabel(f'PC2 ({pca.explained_variance_ratio_[1]*100:.1f}% variance)', fontsize=12)
axes[1].set_title('Kmeans prediction', fontsize=14, fontweight='bold')
Out[165]:
Text(0.5, 1.0, 'Kmeans prediction')
In [166]:
import numpy as np
# Compute covariance matrix
cov_matrix = np.cov(X_scaled.T)
print(cov_matrix)
[[ 1.00671141 -0.11835884 0.87760447 0.82343066] [-0.11835884 1.00671141 -0.43131554 -0.36858315] [ 0.87760447 -0.43131554 1.00671141 0.96932762] [ 0.82343066 -0.36858315 0.96932762 1.00671141]]
In [167]:
# Step 4: Compute eigenvalues and eigenvectors
eigenvalues, eigenvectors = np.linalg.eig(cov_matrix)
print(eigenvalues[0:2]/eigenvalues.sum()*100)
[72.96244541 22.85076179]
In [168]:
from matplotlib.pylab import cm
plt.figure(figsize=(16,2))
sns.heatmap(pca.components_,cmap=cm.seismic,yticklabels=['PC1','PC2'],
square=True, cbar_kws={"orientation": "horizontal"});
Hierarchical clustering¶
Building a tree-like structure - a hierarchy - of clusters, with
- Each data point starts as its own cluster (Agglomerative/Bottom-up) OR all points start as one cluster (Divisive/Top-down)
- Clusters are progressively merged (or split)
- The result is a nested hierarchy showing relationships at all levels
Complete Linkage¶
Average Linkage¶
Ward Linkage¶
In [4]:
import numpy as np
import matplotlib.pyplot as plt
from sklearn import datasets
# Set random seed for reproducibility
np.random.seed(42)
# Generate datasets
def generate_datasets():
"""Generate various datasets similar to the image"""
datasets_list = []
# 1. Concentric circles
noisy_circles = datasets.make_circles(n_samples=300, factor=0.5, noise=0.05)
datasets_list.append(('Circles', noisy_circles[0], noisy_circles[1]))
# 2. Noisy moons
noisy_moons = datasets.make_moons(n_samples=300, noise=0.05)
datasets_list.append(('Moons', noisy_moons[0], noisy_moons[1]))
# 3. Blobs (varied)
blobs = datasets.make_blobs(n_samples=300, centers=3, cluster_std=0.5, random_state=8)
datasets_list.append(('Blobs', blobs[0], blobs[1]))
# 4. Anisotropic blobs
X, y = datasets.make_blobs(n_samples=300, centers=3, random_state=42)
transformation = [[0.6, -0.6], [-0.4, 0.8]]
X_aniso = np.dot(X, transformation)
datasets_list.append(('Anisotropic', X_aniso, y))
# 5. Different variances
varied = datasets.make_blobs(n_samples=300, centers=3,
cluster_std=[1.0, 2.5, 0.5], random_state=42)
datasets_list.append(('Varied', varied[0], varied[1]))
return datasets_list
In [5]:
datasets_list = generate_datasets()
linkage_methods = ['single', 'average', 'complete', 'ward']
fig, axes = plt.subplots(1, len(datasets_list), figsize=(15, 5));
cnt = 0;
for ax in axes:
ax.scatter(datasets_list[cnt][1][:,0], datasets_list[cnt][1][:,1],
color='gray',s=20,alpha=0.7);
ax.set_title(datasets_list[cnt][0], fontsize=14, fontweight='bold')
cnt += 1;
In [6]:
colors = ['#1f77b4', '#ff7f0e', '#2ca02c'] # Blue, Orange, Green
fig, axes = plt.subplots(1, len(datasets_list), figsize=(15, 5));
cnt = 0;
for ax in axes:
for label in datasets_list[cnt][2]:
mask = datasets_list[cnt][2] == label
ax.scatter(datasets_list[cnt][1][mask,0], datasets_list[cnt][1][mask,1], c=colors[label.astype('uint8')], s=20,alpha=0.7);
ax.set_title(datasets_list[cnt][0], fontsize=14, fontweight='bold')
cnt += 1;
In [14]:
# Hierarchical clustering with different linkage methods
def perform_clustering(X, method='single', n_clusters=3):
"""Perform hierarchical clustering"""
# Standardize features
X_scaled = StandardScaler().fit_transform(X)
# Perform hierarchical clustering
start_time = time.time()
Z = linkage(X_scaled, method=method)
elapsed_time = time.time() - start_time
# Get cluster labels
labels = fcluster(Z, n_clusters, criterion='maxclust')
return labels, elapsed_time
In [17]:
from scipy.cluster.hierarchy import dendrogram, linkage
from scipy.cluster.hierarchy import fcluster
from sklearn.preprocessing import StandardScaler
import time
linkage_methods = ['single', 'average', 'complete', 'ward']
fig, axes = plt.subplots(len(datasets_list), len(linkage_methods),
figsize=(16, 18))
# Color map
colors = ['#1f77b4', '#ff7f0e', '#2ca02c'] # Blue, Orange, Green
true_clusters=[2,2,3,3,3,1];
cnt=0;
for i, (dataset_name, X, y_true) in enumerate(datasets_list):
for j, method in enumerate(linkage_methods):
ax = axes[i, j]
# Perform clustering
labels, elapsed_time = perform_clustering(X, method=method, n_clusters=true_clusters[cnt])
# Plot the results
for cluster_id in range(1, 4):
mask = labels == cluster_id
ax.scatter(X[mask, 0], X[mask, 1],
c=colors[cluster_id-1], s=20, alpha=0.7,
edgecolors='none')
# Remove axes
ax.set_xticks([])
ax.set_yticks([])
# Add column titles
if i == 0:
title = method.replace('_', ' ').title() + ' Linkage'
ax.set_title(title, fontsize=14, fontweight='bold')
# Add row labels
if j == 0:
ax.set_ylabel(dataset_name, fontsize=12, rotation=0,
ha='right', va='center', labelpad=30)
cnt += 1;
plt.tight_layout()
plt.show()
In [169]:
from scipy.cluster.hierarchy import linkage, fcluster, dendrogram
# 1. Load and standardize IRIS data
iris = load_iris()
X = iris.data
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
# 2. Perform hierarchical clustering
Z = linkage(X_scaled, method='ward')
# 3. Cut dendrogram to get 3 clusters
clusters = fcluster(Z, t=3, criterion='maxclust') - 1
# 4. Visualize dendrogram
fig, ax = plt.subplots(figsize=(12, 6))
# Create dendrogram
dendrogram(Z, ax=ax,
labels=y, # Color by true species
color_threshold=0,
above_threshold_color='gray')
ax.set_title('Hierarchical Clustering Dendrogram (Ward Linkage)',
fontsize=14, fontweight='bold')
ax.set_xlabel('Sample Index', fontsize=12)
ax.set_ylabel('Distance (Ward)', fontsize=12)
# Add horizontal line showing where we cut for k=3
ax.axhline(y=6, color='red', linestyle='--', linewidth=2,
label='Cut at k=3 clusters')
ax.legend(fontsize=10)
plt.show()
print(f"Created {len(set(clusters))} clusters")
print(f"Cluster sizes: {[sum(clusters == i) for i in range(3)]}")
Created 3 clusters Cluster sizes: [np.int64(49), np.int64(30), np.int64(71)]
PCA vs LDA¶
Now let us consider data on a manifold¶
In [21]:
from sklearn.datasets import make_swiss_roll
import matplotlib.pyplot as plt
points, colors = make_swiss_roll(n_samples=3000, random_state=10);
In [22]:
#Let's plot the Swiss roll
fig = plt.figure(figsize=(6, 6))
ax = fig.add_subplot(111, projection="3d")
fig.add_axes(ax)
ax.scatter(
points[:, 0], points[:, 1], points[:, 2], c=colors,
s=50, alpha=0.5, cmap=plt.cm.Spectral
)
ax.set_title("Swiss Roll")
ax.view_init(azim=-66, elev=10)
#_ = ax.text2D(0.8, 0.05, s="n_samples=1500", transform=ax.transAxes)
In [23]:
#lets perform PCA on the swiss roll
from sklearn.decomposition import PCA
pca_model = PCA(n_components=2)
pca_result = pca_model.fit_transform(points)
plt.figure(figsize=(6, 6))
plt.scatter(pca_result[:, 0], pca_result[:, 1], c=colors,
s=50, alpha=0.5, cmap=plt.cm.Spectral)
plt.xlabel('First Principal Component')
plt.ylabel('Second Principal Component')
plt.title('PCA of Swiss Roll')
plt.grid(True)
plt.tight_layout()
plt.show()
In [24]:
# Is the following better 2D projection though?
In [25]:
plt.figure(figsize=(6, 6))
plt.scatter(colors,points[:,1], c=colors, s=50, alpha=0.5, cmap=plt.cm.Spectral)
plt.title('Unwrapping the Swissroll')
plt.grid(True)
plt.tight_layout()
plt.show()
In [27]:
import umap
umap_model = umap.UMAP()
umap_result = umap_model.fit_transform(points, n_neighbors=100, min_dist = .5)
plt.figure(figsize=(6, 6))
plt.scatter(umap_result[:, 0], umap_result[:, 1], c=colors, s=50, alpha=0.5, cmap=plt.cm.Spectral)
plt.xlabel('UMAP 1')
plt.ylabel('UMAP 2')
plt.title('UMAP of Swiss Roll')
plt.grid(True)
plt.tight_layout()
plt.show();
UMAP: Uniform Manifold Approximation & Projection¶
https://umap-learn.readthedocs.io/en/latest/index.html
Idea: Prioritize respecting local distances in embedding space.
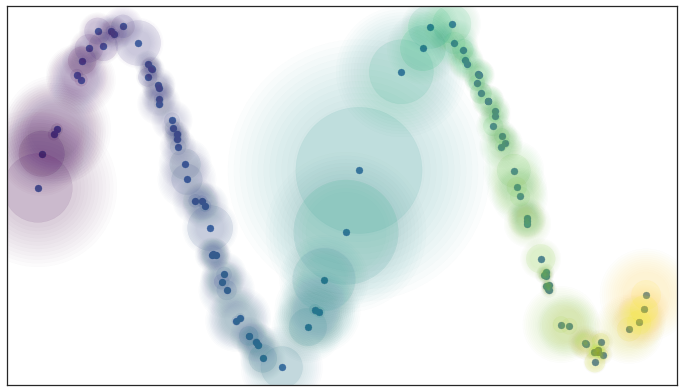
UMAP: Uniform Manifold Approximation & Projection¶
UMAP constructs a $k$-nearest neighbors graph of the data. It uses an approximate stochastic algorithm to do this quickly.
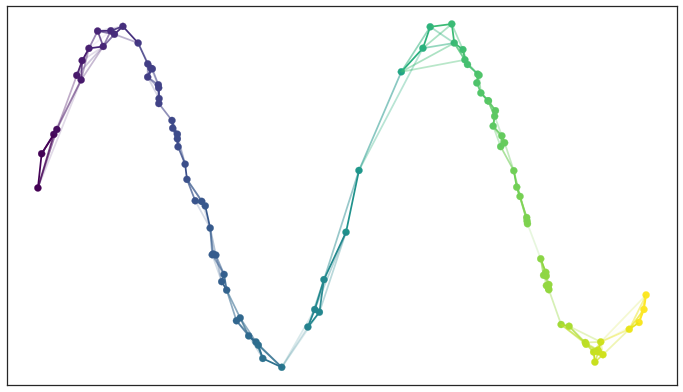
UMAP Parameters¶
n_neighbors(15): Number of nearest neighbors to consider. Increase to respect global structure more.min_dist(0.1): Minimum distance between points in reduced space. Decrease to have more tightly clustered points.metric("euclidean"): How distance is computed. Can provide user-supplied functions in addition to standard metrics
In [28]:
import numpy as np
import matplotlib.pyplot as plt
from sklearn.datasets import make_moons
from umap import UMAP
# Generate synthetic data
n_samples = 1000
X, y = make_moons(n_samples=n_samples, noise=0.1, random_state=42)
plt.figure(figsize=(6, 6))
plt.scatter(X[:, 0], X[:, 1], c=y, s=10, alpha=0.5, cmap=plt.cm.Spectral)
Out[28]:
<matplotlib.collections.PathCollection at 0x7a7e531f0690>
In [30]:
# Set up the plot
fig, axs = plt.subplots(3, 3, figsize=(15, 15))
fig.suptitle("UMAP Parameter Sensitivity", fontsize=16)
# Define parameter ranges
n_neighbors_range = [5, 15, 50]
min_dist_range = [0.0, 0.1, 0.5]
# Generate UMAP projections for different parameter combinations
for i, n_neighbors in enumerate(n_neighbors_range):
for j, min_dist in enumerate(min_dist_range):
umap = UMAP(n_neighbors=n_neighbors, min_dist=min_dist, random_state=42, n_jobs=1)
X_umap = umap.fit_transform(X)
axs[i, j].scatter(X_umap[:, 0], X_umap[:, 1], c=y, cmap=plt.cm.Spectral, s=5)
axs[i, j].set_title(f'n_neighbors={n_neighbors}, min_dist={min_dist}')
axs[i, j].set_xticks([])
axs[i, j].set_yticks([])
plt.tight_layout()
plt.show()
Next time¶
-RNA seq